Molecular Network Of Peptide Filtered Lc Ms Ms Datasets Of Um Botanical Download Scientific

Molecular Network Of Peptide-filtered LC-MS/MS Datasets Of UM Botanical... | Download Scientific ...
Molecular Network Of Peptide-filtered LC-MS/MS Datasets Of UM Botanical... | Download Scientific ... Herein, we demonstrate that the lancl enzymes can remove gsh adducts from c glutathionylated peptides with dl or ll lanthionine stereochemistry. these two advances will aid synthetic. To this end, a new approach integrating mixed mode cationic exchange based solid phase extraction (mcx spe) coupled with liquid chromatography mass spectrometry (lc–ms) and feature based molecular networking (fbmn) was developed.

Molecular Network Of Peptide-filtered LC-MS/MS Datasets Of UM Botanical... | Download Scientific ...
Molecular Network Of Peptide-filtered LC-MS/MS Datasets Of UM Botanical... | Download Scientific ... Nist is developing a peptide mass spectral library as an extension of the nist/epa/nih mass spectral library. the purpose of the library is to provide peptide reference data for laboratories employing mass spectrometry based proteomics methods for protein analysis. These new approaches, including recurrent neural networks and convolutional neural networks, use predicted in silico spectral libraries rather than experimental libraries to achieve higher. In this work, we have created a new analysis tool, phosfox, for processing and comparing phosphoproteomic data from multiple samples and several different database search algorithms. Deep learning models for prediction of three key lc ms/ms properties from peptide sequences were developed. the lc ms/ms properties or behaviors are indexed retention times (irt), ms1 or survey scan charge state distributions, and sequence ion intensities of hcd spectra.

Molecular Network Of Peptide-filtered LC-MS/MS Datasets Of UM Botanical... | Download Scientific ...
Molecular Network Of Peptide-filtered LC-MS/MS Datasets Of UM Botanical... | Download Scientific ... In this work, we have created a new analysis tool, phosfox, for processing and comparing phosphoproteomic data from multiple samples and several different database search algorithms. Deep learning models for prediction of three key lc ms/ms properties from peptide sequences were developed. the lc ms/ms properties or behaviors are indexed retention times (irt), ms1 or survey scan charge state distributions, and sequence ion intensities of hcd spectra. Gnps is a web based mass spectrometry ecosystem that aims to be an open access knowledge base for community wide organization and sharing of raw, processed, or annotated fragmentation mass spectrometry data (ms/ms). This paper introduces an automated noise filtering method based on the construction of orthogonal polynomials. by subdividing the spectrum into a variable number (3 to 11) of bins, peaks that are considered "noise" are identified at a local level. Utilizing a pre generated plant extract library, we subjected botanicals to lc ms/ms based molecular networking to determine their chemical composition and relatively quantify already known metabolites. Download scientific diagram | molecular family from the molecular network of lc ms/ms datasets from 14 extracts of heterologous e. coli expressing agpi rendered by gnps (47).

Molecular Family From The Molecular Network Of LC-MS/MS Datasets From... | Download Scientific ...
Molecular Family From The Molecular Network Of LC-MS/MS Datasets From... | Download Scientific ... Gnps is a web based mass spectrometry ecosystem that aims to be an open access knowledge base for community wide organization and sharing of raw, processed, or annotated fragmentation mass spectrometry data (ms/ms). This paper introduces an automated noise filtering method based on the construction of orthogonal polynomials. by subdividing the spectrum into a variable number (3 to 11) of bins, peaks that are considered "noise" are identified at a local level. Utilizing a pre generated plant extract library, we subjected botanicals to lc ms/ms based molecular networking to determine their chemical composition and relatively quantify already known metabolites. Download scientific diagram | molecular family from the molecular network of lc ms/ms datasets from 14 extracts of heterologous e. coli expressing agpi rendered by gnps (47).
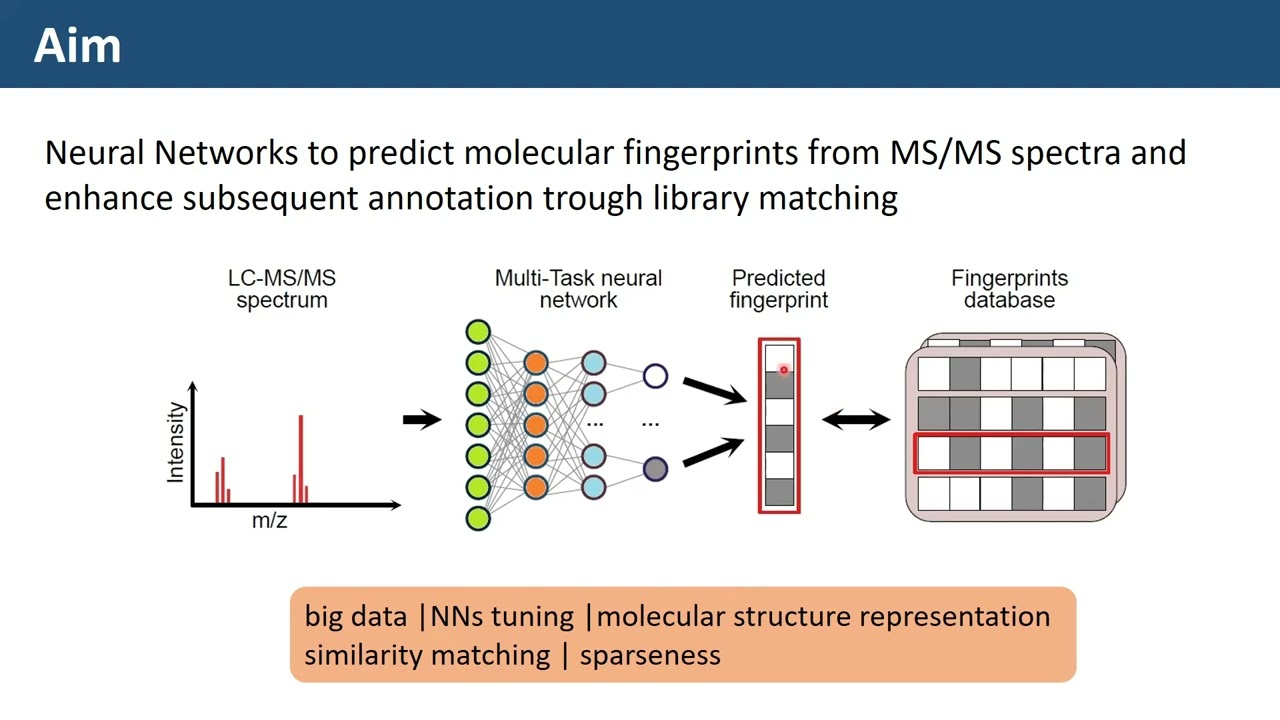
Monday webinar - Chemometrics and molecular fingerprints to enhance LC-MS/MS spectral match
Monday webinar - Chemometrics and molecular fingerprints to enhance LC-MS/MS spectral match
Related image with molecular network of peptide filtered lc ms ms datasets of um botanical download scientific
Related image with molecular network of peptide filtered lc ms ms datasets of um botanical download scientific
About "Molecular Network Of Peptide Filtered Lc Ms Ms Datasets Of Um Botanical Download Scientific"
















Comments are closed.