Mqss 2018 L20 Peptide Ms Ms Spectrum Prediction Using Deep Learning Peter Cimermancic

Peptide MS/MS Spectrum And IRT Prediction A A Graphical Illustration Of... | Download Scientific ...
Peptide MS/MS Spectrum And IRT Prediction A A Graphical Illustration Of... | Download Scientific ... Full title: high quality peptide ms/ms spectrum prediction using deep learning and its application in dia data analysismqss website: http://summerschool.maxq. In this paper, we present a deep learning approach that can predict the complete spectra (both backbone and nonbackbone ions) directly from peptide sequences. we made no assumptions or expectations on which kind of ions to predict but instead predicting the intensities for all possible m / z.

Deep Learning Tool For Predicting MS/MS Spectrum | Download Scientific Diagram
Deep Learning Tool For Predicting MS/MS Spectrum | Download Scientific Diagram Here, we present pdeep, a deep neural network based model for the spectrum prediction of peptides. In this manuscript, we present deepmspeptide, a bioinformatic tool that uses a deep learning method to predict proteotypic peptides exclusively based on the peptide amino acid sequences. deepmspeptide is available at https://github.com/vsegurar/deepmspeptide. supplementary data are available at bioinformatics online. Longstanding methods for peptide identification, such as search engines and experimental spectral libraries, are being superseded by deep learning models that allow the fragmentation spectra. To overcome this challenge, we have developed deepms, a deep learning based spectra identification algorithm that overcomes the speed limitations of traditional spectra identification methods. we conducted comprehensive benchmark tests, comparing six deep learning algorithms.

Deep Learning Tool For Predicting MS/MS Spectrum | Download Scientific Diagram
Deep Learning Tool For Predicting MS/MS Spectrum | Download Scientific Diagram Longstanding methods for peptide identification, such as search engines and experimental spectral libraries, are being superseded by deep learning models that allow the fragmentation spectra. To overcome this challenge, we have developed deepms, a deep learning based spectra identification algorithm that overcomes the speed limitations of traditional spectra identification methods. we conducted comprehensive benchmark tests, comparing six deep learning algorithms. In this review, we will follow the workflow of a proteomic ms experiment to demonstrate how ai is applied in current prediction models. In this paper, we present a deep learning approach that can predict the complete spectra (both backbone and non backbone ions) directly from peptide sequences. we made no assumptions or expectations on which kind of ions to predict but instead predicting the intensities for all possible m/z. Here, we demonstrate that machine learning can predict peptide fragmentation patterns in mass spectrometers with accuracy within the uncertainty of measurement. moreover, analysis of. Here we introduce pdeepxl, a deep neural network to predict ms/ms spectra of cross linked peptide pairs. to train pdeepxl, we used the transfer learning technique because it facilitated the training with limited benchmark data of cross linked peptide pairs.

How To Train Your Modified Peptide MS/MS Spectrum Predictor? - PROTrEIN
How To Train Your Modified Peptide MS/MS Spectrum Predictor? - PROTrEIN In this review, we will follow the workflow of a proteomic ms experiment to demonstrate how ai is applied in current prediction models. In this paper, we present a deep learning approach that can predict the complete spectra (both backbone and non backbone ions) directly from peptide sequences. we made no assumptions or expectations on which kind of ions to predict but instead predicting the intensities for all possible m/z. Here, we demonstrate that machine learning can predict peptide fragmentation patterns in mass spectrometers with accuracy within the uncertainty of measurement. moreover, analysis of. Here we introduce pdeepxl, a deep neural network to predict ms/ms spectra of cross linked peptide pairs. to train pdeepxl, we used the transfer learning technique because it facilitated the training with limited benchmark data of cross linked peptide pairs.

How To Train Your Modified Peptide MS/MS Spectrum Predictor? - PROTrEIN
How To Train Your Modified Peptide MS/MS Spectrum Predictor? - PROTrEIN Here, we demonstrate that machine learning can predict peptide fragmentation patterns in mass spectrometers with accuracy within the uncertainty of measurement. moreover, analysis of. Here we introduce pdeepxl, a deep neural network to predict ms/ms spectra of cross linked peptide pairs. to train pdeepxl, we used the transfer learning technique because it facilitated the training with limited benchmark data of cross linked peptide pairs.

Figure 1 From Deep Learning-Enabled MS/MS Spectrum Prediction Facilitates Automated ...
Figure 1 From Deep Learning-Enabled MS/MS Spectrum Prediction Facilitates Automated ...
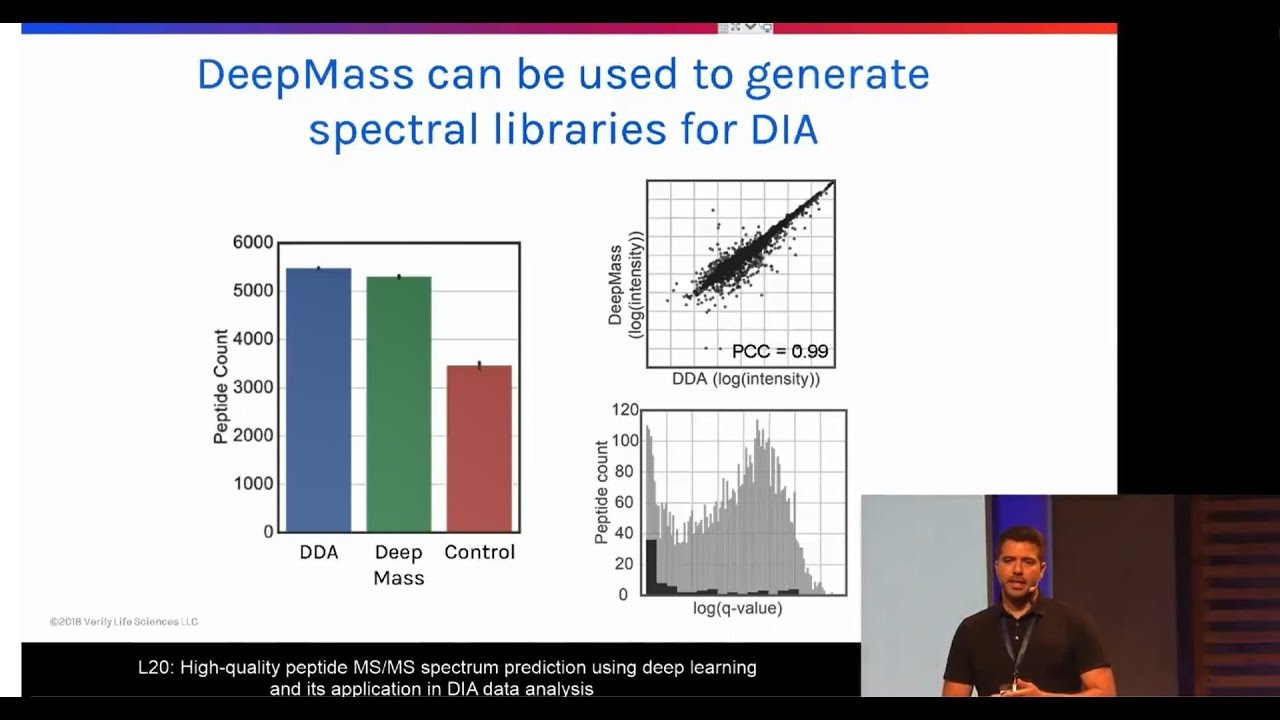
MQSS 2018 | L20: Peptide MS/MS spectrum prediction using deep learning | Peter Cimermancic
MQSS 2018 | L20: Peptide MS/MS spectrum prediction using deep learning | Peter Cimermancic
Related image with mqss 2018 l20 peptide ms ms spectrum prediction using deep learning peter cimermancic
Related image with mqss 2018 l20 peptide ms ms spectrum prediction using deep learning peter cimermancic
About "Mqss 2018 L20 Peptide Ms Ms Spectrum Prediction Using Deep Learning Peter Cimermancic"
















Comments are closed.