Multi Omics Studies With Histopathology Using Deep Learning Biorender Science Templates

Multi-omics Studies With Histopathology Using Deep Learning | BioRender Science Templates
Multi-omics Studies With Histopathology Using Deep Learning | BioRender Science Templates Customize this multi omics studies with histopathology using deep learning template with biorender. create professional, scientifically accurate visuals in minutes. Here we present histoplexer, a deep learning framework that generates spatially resolved protein multiplexes directly from standard haematoxylin and eosin (h&e) histopathology images.

(PDF) Deep Learning Approaches In Histopathology
(PDF) Deep Learning Approaches In Histopathology This paper proposes a deep learning method to jointly model multi omics and histopathology slides. the model can handle incomplete data (e.g. patients might be missing different modalities) and aims to shed light on the activity of different pathways in different cancer types. We sought to determine which complex models of large scale multi omics datasets could be rendered human interpretable using human intuition in the absence of attention maps to enhance uptake of this technology in translational research and clinical practice. Through both single task and multi task learning frameworks, we demonstrate that pathway and mirna predictions not only enhance interpretability but also provide valuable prognostic insights. Ai, utilizing its sophisticated machine learning algorithms and deep learning frameworks, provides the computational power and expertise required to analyze the intricacies of multi omics data.

Figure 1 From Deep Learning Approaches In Histopathology | Semantic Scholar
Figure 1 From Deep Learning Approaches In Histopathology | Semantic Scholar Through both single task and multi task learning frameworks, we demonstrate that pathway and mirna predictions not only enhance interpretability but also provide valuable prognostic insights. Ai, utilizing its sophisticated machine learning algorithms and deep learning frameworks, provides the computational power and expertise required to analyze the intricacies of multi omics data. Here, we present a systematic survey of the academic literature from 2010 to november 2023, aiming to quantify the application of dl for pathology, genomics, and the combined use of both data types. We identified a total of 11 studies meeting the inclusion criteria, namely studies that used convolutional neural networks for haematoxylin and eosin image analysis of patients with cancer in combination with integrated omics data. This study investigates whether deep learning (dl), a series of advanced computer techniques, can perform molecular profiling directly from routinely collected images of tumor specimens used for diagnostic purposes. This thesis tackles the development of computational models and learning strategies adept at deciphering complex, high dimensional interactions. a significant focus is also placed on the explainability of these ai driven models, ensuring that predictions are understandable and clinically actionable.

Exploring Regions Of Interest: Visualizing Histological Image Classification For Breast Cancer ...
Exploring Regions Of Interest: Visualizing Histological Image Classification For Breast Cancer ... Here, we present a systematic survey of the academic literature from 2010 to november 2023, aiming to quantify the application of dl for pathology, genomics, and the combined use of both data types. We identified a total of 11 studies meeting the inclusion criteria, namely studies that used convolutional neural networks for haematoxylin and eosin image analysis of patients with cancer in combination with integrated omics data. This study investigates whether deep learning (dl), a series of advanced computer techniques, can perform molecular profiling directly from routinely collected images of tumor specimens used for diagnostic purposes. This thesis tackles the development of computational models and learning strategies adept at deciphering complex, high dimensional interactions. a significant focus is also placed on the explainability of these ai driven models, ensuring that predictions are understandable and clinically actionable.
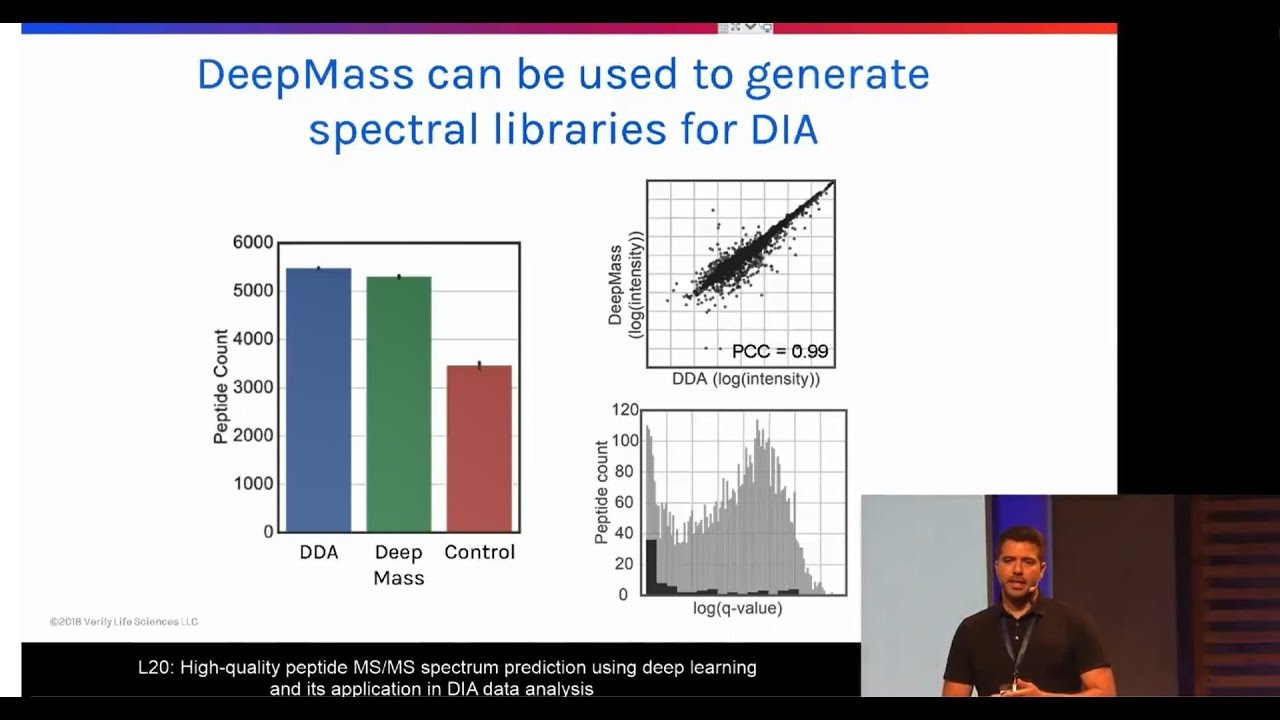
MQSS 2018 | L20: Peptide MS/MS spectrum prediction using deep learning | Peter Cimermancic
MQSS 2018 | L20: Peptide MS/MS spectrum prediction using deep learning | Peter Cimermancic
Related image with multi omics studies with histopathology using deep learning biorender science templates
Related image with multi omics studies with histopathology using deep learning biorender science templates
About "Multi Omics Studies With Histopathology Using Deep Learning Biorender Science Templates"
















Comments are closed.