Pdf Mass Spectrometry Based Bottom Up Proteomics Sample Preparation Lc Ms Ms Analysis And

Proteomes | Free Full-Text | Selecting Sample Preparation Workflows For Mass Spectrometry-Based ...
Proteomes | Free Full-Text | Selecting Sample Preparation Workflows For Mass Spectrometry-Based ... The workflow described herein details sample preparation from tissues through protein extraction, proteolysis, and purification to generate peptides for ms analysis. Here, we provide a comprehensive overview of different proteomics methods to aid the novice and experienced researcher. we cover from biochemistry basics and protein extraction to biological interpretation and orthogonal validation.

(PDF) Mass Spectrometry-Based Bottom-Up Proteomics: Sample Preparation, LC-MS/MS Analysis, And ...
(PDF) Mass Spectrometry-Based Bottom-Up Proteomics: Sample Preparation, LC-MS/MS Analysis, And ... This article outlines the steps required to prepare peptide samples for ms within the context of a bottom up proteomics workflow. the bottom up proteomics methods described are versatile and can be used for discovery and targeted analyses in qualitative and quantitative workflows. We performed analysis of dynamic range, sensitivity, and robustness using our microflow lc nanospray ms platform and compared the results to those obtained from microflow lc ms and nanolc ms, respectively. Simplified conceptual overview of a typical untargeted bottom up mass spectrometry based proteomic experiment. using a protease, proteins are digested into peptides, which are separated by liquid chromatography, sprayed into a mass spectrometer, and enter the gas phase as ions. However, by far the most common method for proteomics is based on mass spectrometry coupled to liquid chromatography (lc). modern proteomics had its roots in the early 1980s with the analysis of peptides by mass spectrometry and low efficiency ion sources.

(PDF) Bottom-Up Proteomics: Advancements In Sample Preparation
(PDF) Bottom-Up Proteomics: Advancements In Sample Preparation Simplified conceptual overview of a typical untargeted bottom up mass spectrometry based proteomic experiment. using a protease, proteins are digested into peptides, which are separated by liquid chromatography, sprayed into a mass spectrometer, and enter the gas phase as ions. However, by far the most common method for proteomics is based on mass spectrometry coupled to liquid chromatography (lc). modern proteomics had its roots in the early 1980s with the analysis of peptides by mass spectrometry and low efficiency ion sources. In this review, we summarize the current state of mass spectrometry applied to bottom up proteomics, the approach that focuses on analyzing peptides obtained from proteolytic digestion of proteins. Liquid chromatography–tandem mass spectrometry (lc–ms/ms) based proteomics is a powerful technique for profiling proteomes of cells, tissues, and body fluids. typical bottom up proteomic workflows consist of the following three major steps: sample preparation, lc–ms/ms analysis, and data analysis. The workflow described herein details sample preparation from tissues through protein extraction, proteolysis, and purification to generate peptides for ms analysis.

(PDF) Facile Preparation Of Peptides For Mass Spectrometry Analysis In Bottom‐Up Proteomics ...
(PDF) Facile Preparation Of Peptides For Mass Spectrometry Analysis In Bottom‐Up Proteomics ... In this review, we summarize the current state of mass spectrometry applied to bottom up proteomics, the approach that focuses on analyzing peptides obtained from proteolytic digestion of proteins. Liquid chromatography–tandem mass spectrometry (lc–ms/ms) based proteomics is a powerful technique for profiling proteomes of cells, tissues, and body fluids. typical bottom up proteomic workflows consist of the following three major steps: sample preparation, lc–ms/ms analysis, and data analysis. The workflow described herein details sample preparation from tissues through protein extraction, proteolysis, and purification to generate peptides for ms analysis.

(PDF) Application Of Mass Spectrometry-Based Proteomics To Barley Research
(PDF) Application Of Mass Spectrometry-Based Proteomics To Barley Research The workflow described herein details sample preparation from tissues through protein extraction, proteolysis, and purification to generate peptides for ms analysis.
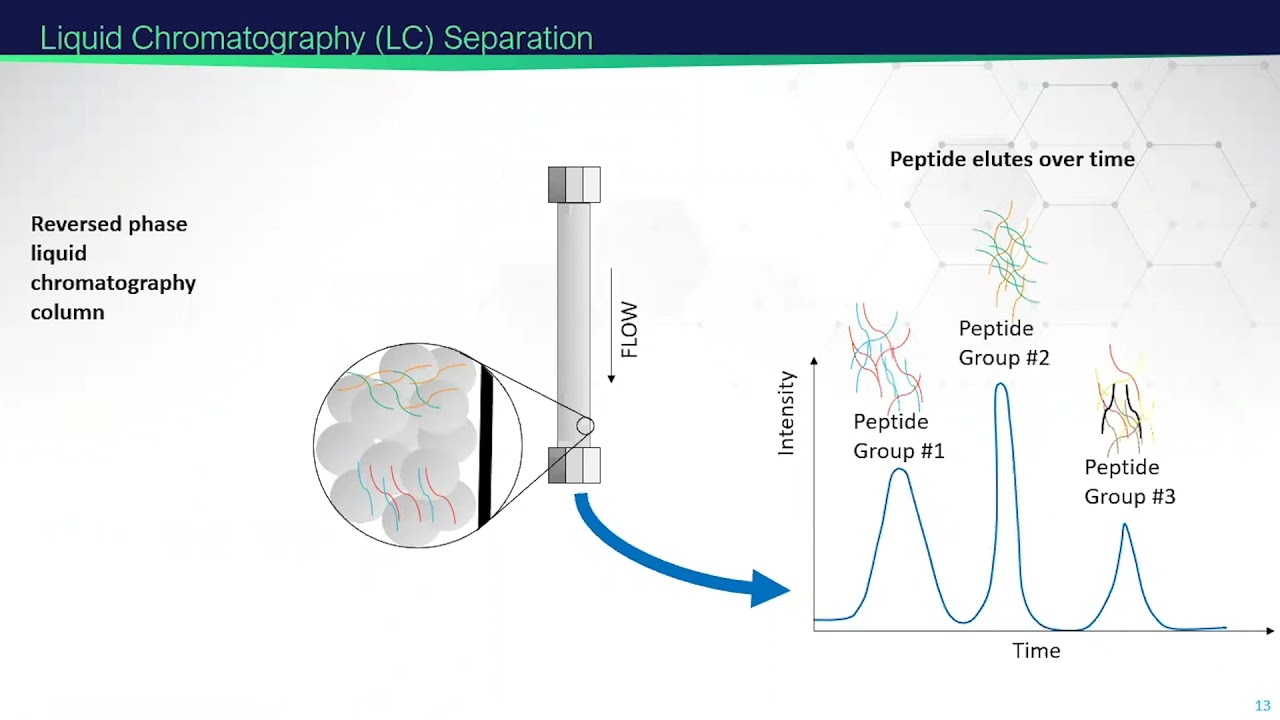
Mass Spectrometry-Based Proteomics | 2021 EMSL Summer School
Mass Spectrometry-Based Proteomics | 2021 EMSL Summer School
Related image with pdf mass spectrometry based bottom up proteomics sample preparation lc ms ms analysis and
Related image with pdf mass spectrometry based bottom up proteomics sample preparation lc ms ms analysis and
About "Pdf Mass Spectrometry Based Bottom Up Proteomics Sample Preparation Lc Ms Ms Analysis And"
















Comments are closed.